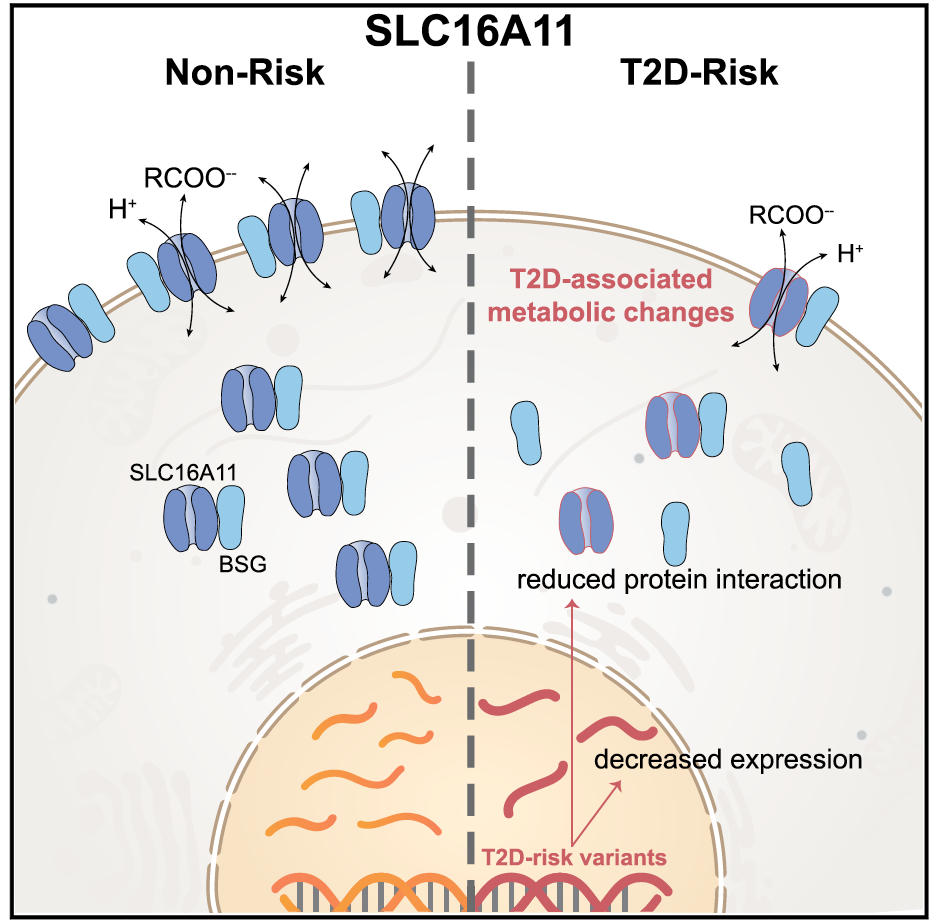
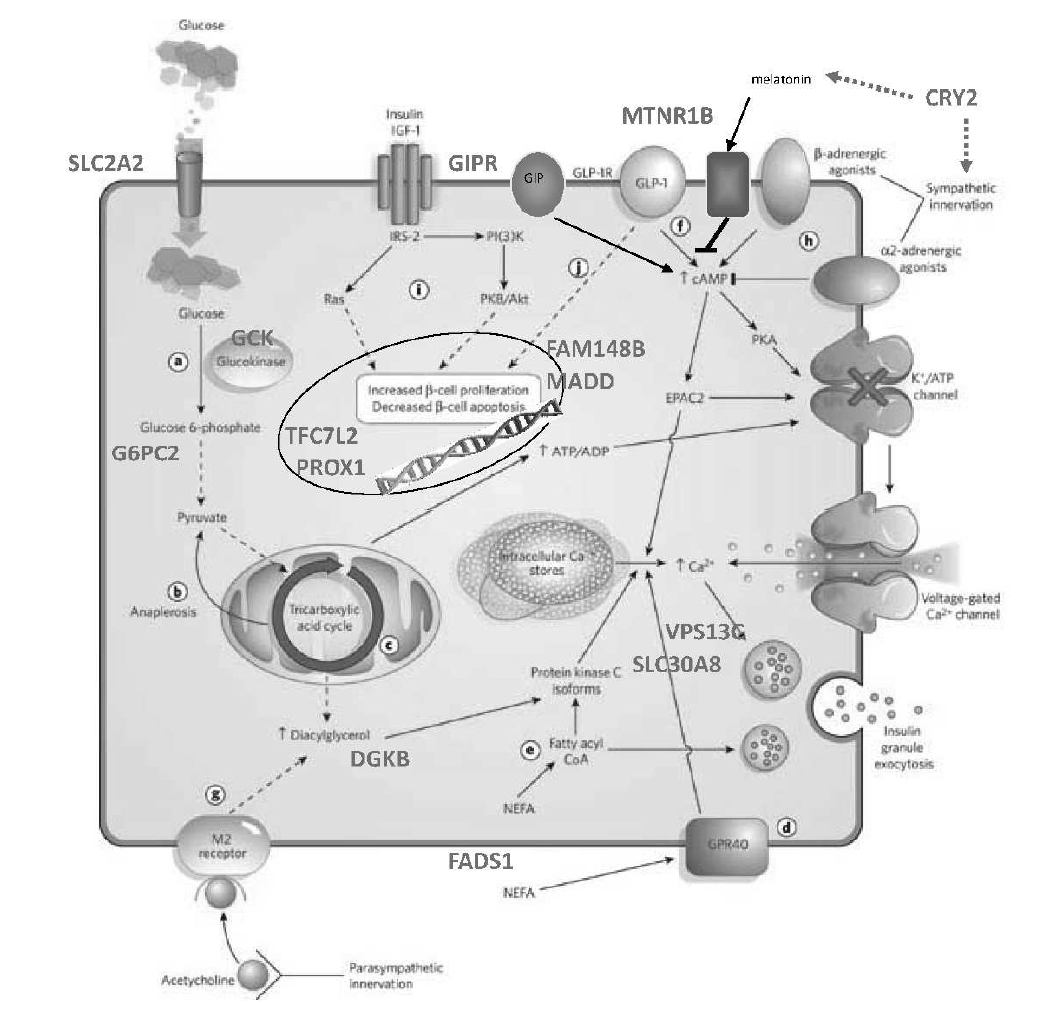
We have entered a new era in which technological developments coupled with expanded computational power and increased statistical sophistication have enabled the global query of discrete biological axes. Manufacturing advances introduced arrays that could measure mRNA transcript levels or DNA single nucleotide variation comprehensively in a single experiment. A deeper knowledge of the patterns of human genetic variation allowed for the explosion of genome-wide association studies (GWAS). Remarkable efficiency gains in high-throughput next-generation sequencing seeded whole-exome and whole-genome sequencing studies, as well as ancillary explorations of the transcriptome (RNAseq), transcription factor binding sites (ChiPseq), open chromatin (ATAC-seq), the epigenome, or the microbiome. Mass spectroscopy can be applied to the study of small metabolites or proteins in organic fluids, including post-translational modifications. In this manner, multiple dimensions of the molecular architecture of biological systems can be interrogated with respect to native and perturbed metabolic states. This technological progress has been accompanied by concomitant enhancements in bioinformatic and analytical tools, often shared publicly in the pre-competitive space.
Crucially, nowadays all of these advances can increasingly be deployed in the organism of interest, the human. Health care systems have digitalized clinical information, and increasingly made the electronic medical record available to clinical investigation. Large private and even national biobanks have been created to streamline this research function, and both funding bodies and scientific journals have required data sharing in central repositories as a condition for research support or publication. We therefore live in the midst of a revolution of big data across all domains of the human experience, ranging from the molecular to the societal dimensions. We practice medicine and conduct research within an unprecedented whirlwind of data, spanning from populations to the individual. It will soon be possible to capture the metabolic state of a single patient at the molecular and cellular levels with great precision through multiple time points in his/her development.
An outstanding but crucial challenge to the field is our ability to integrate these disparate data sources in a manner that informs a holistic view of an organism, such that synergy begets understanding. While genomic explorations have only explained a minor fraction of the genetic contribution to the phenotype, in conjunction with physiological measures they can be used to improve our nosology of the disease, and begin to characterize the clusters that may define specific subtypes.
The integration of physiologic and pharmacogenetic information with genetic discoveries can offer additional insight. By perturbing a live human with a drug that targets a given gene and assessing the response to the perturbation, one may be able to “close the loop” and demonstrate that a gene associated with disease is indeed involved in producing the phenotype of interest. Conversely, drugs that modulate a specific limb of the glucose homeostatic system (insulin secretion, central or peripheral insulin sensitivity), if shown to have differential responses depending on genotype, may serve to prioritize genes in a given associated region.
The Florez lab works with longitudinal observational cohorts (e.g. Framingham Heart Study, the CHARGE Consortium, SEARCH, CAMP), richly phenotyped clinical trials (e.g. the Diabetes Prevention Program, Look AHEAD, TODAY), healthcare biobanks (Partners Biobank, UK Biobank), or our own pharmacogenetic or nutrigenetic studies (SUGAR-MGH, SIGMA) to study genotype-phenotype correlations and analyze physiological measurements to link genetic variation to human organismal biology.