Genetics of Type 2 Diabetes
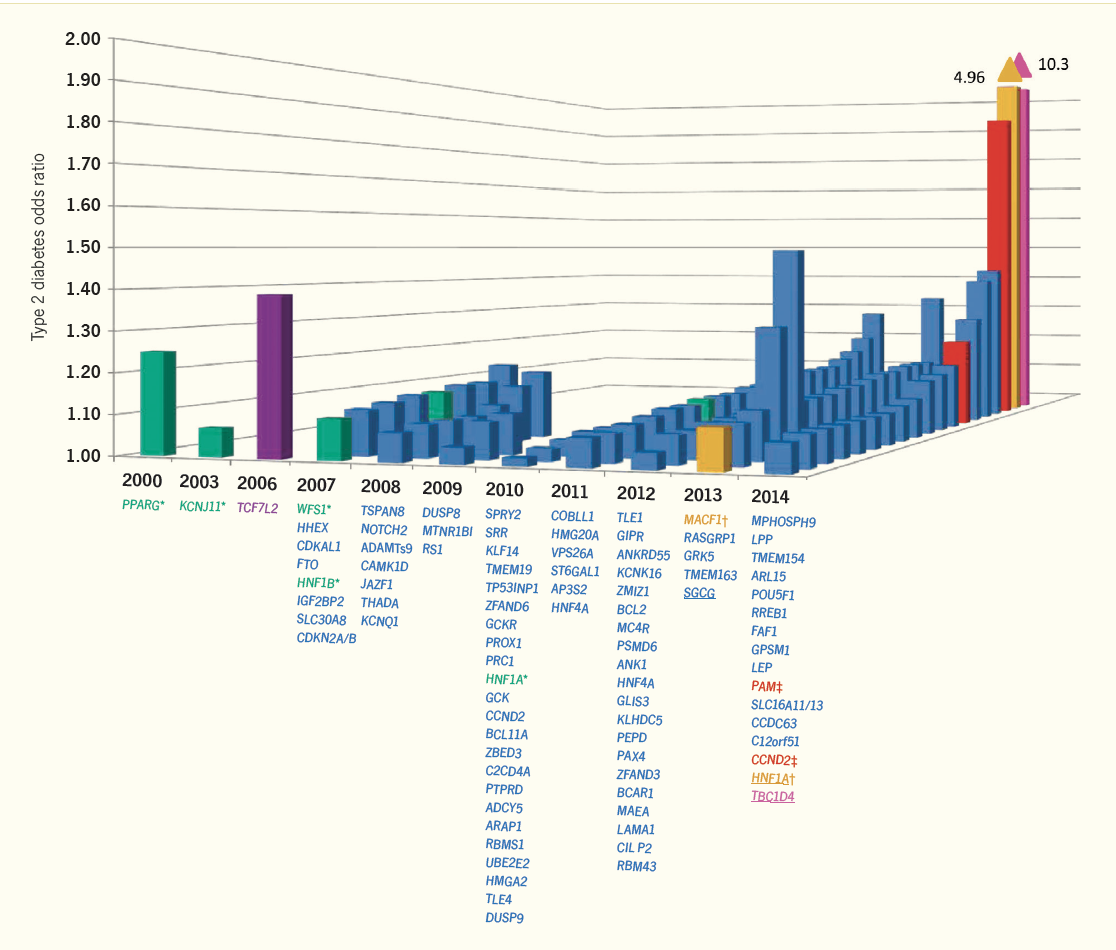
Type 2 diabetes is thought to result from a combination of environmental, behavioral, and genetic factors, with the heritability of type 2 diabetes estimated to be in the range of 25% to 72% based on family and twin studies. Since early 2007, genomewide association studies (GWAS) have led to an explosion of data for the genetics of type 2 diabetes and related traits. These GWAS have occurred on the background of genotyping arrays populated by common single nucleotide polymorphisms (SNPs), deployed in various cohorts that have coalesced to form large international consortia. As a result, a list of genetic loci that influence type 2 diabetes and quantitative glycemic traits has begun to accumulate. Over 100 type 2 diabetes-associated loci have been identified, in addition to others involved in determining quantitative glycemic traits, such as insulin resistance. However, no variant that is widely shared across populations has been found to have a stronger effect than the rs7903146 SNP in TCF7L2, which itself has only a modest effect (odds ratio ~1.4). Nonetheless, GWAS findings have illustrated novel pathways, pointed toward fundamental biology, drawn attention to the role of beta cell dysfunction in type 2 diabetes, confirmed prior epidemiologic observations, and provided possible targets for pharmacotherapy and pharmacogenetic clinical trials.
On the other hand, the causal variants have only been identified for a handful of these loci, a substantial proportion of the heritability of these phenotypes remains unexplained, and this has tempered expectations with regard to their use in clinical prediction. Together, the approximately 100 loci associated with type 2 diabetes thus far explain ~10%–15% of the genetic predisposition to the disease. Limitations of early GWAS included insufficient sample sizes to detect small effects, a nearly exclusive focus on populations of European descent, an imperfect capture of uncommon genetic variants, an incomplete ascertainment of alternate (non-SNP) forms of genetic variation, and the lack of exploration of additional genetic models.
As the community embraces complementary approaches that include systematic fine-mapping, custom-made replication,
denser genotyping arrays, platforms that focus on functional variation, next-generation sequencing techniques, systems biology approaches, and expansion to non-European populations, the coming years will witness exponential growth in the understanding of the genetic architecture of metabolic phenotypes. Whether these findings prove useful in disease prediction or therapeutic decision-making must be tested in rigorously designed clinical trials.



