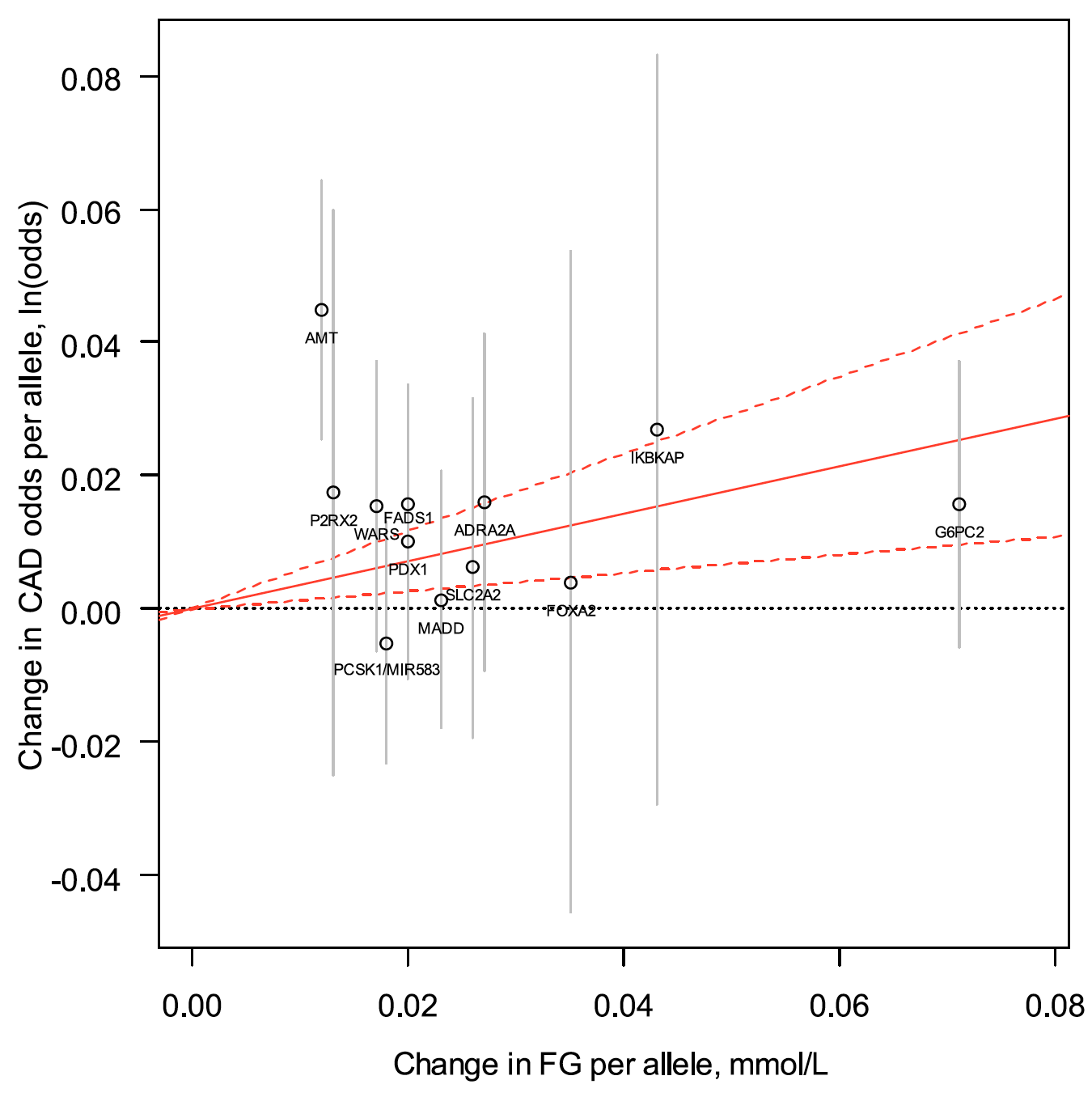
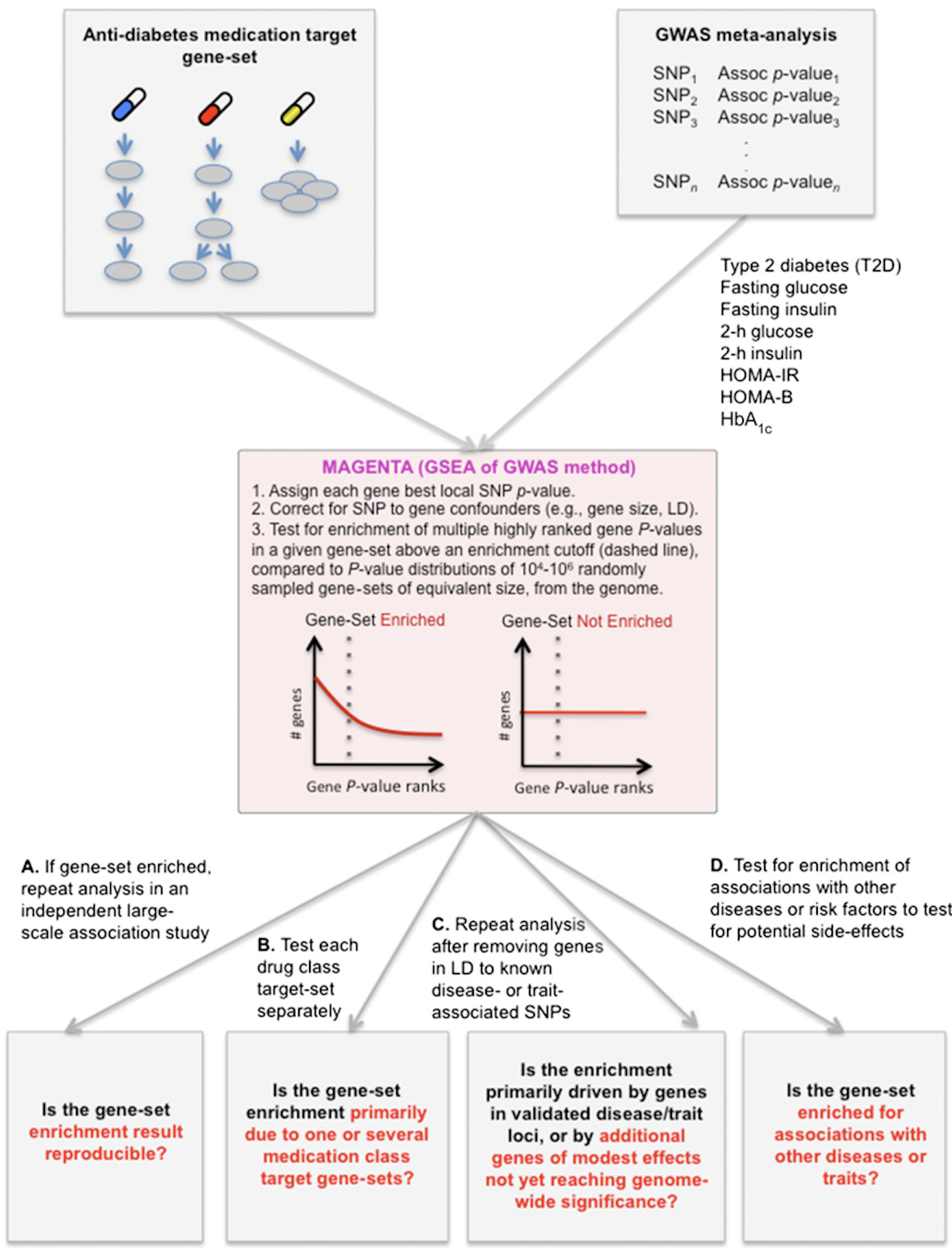
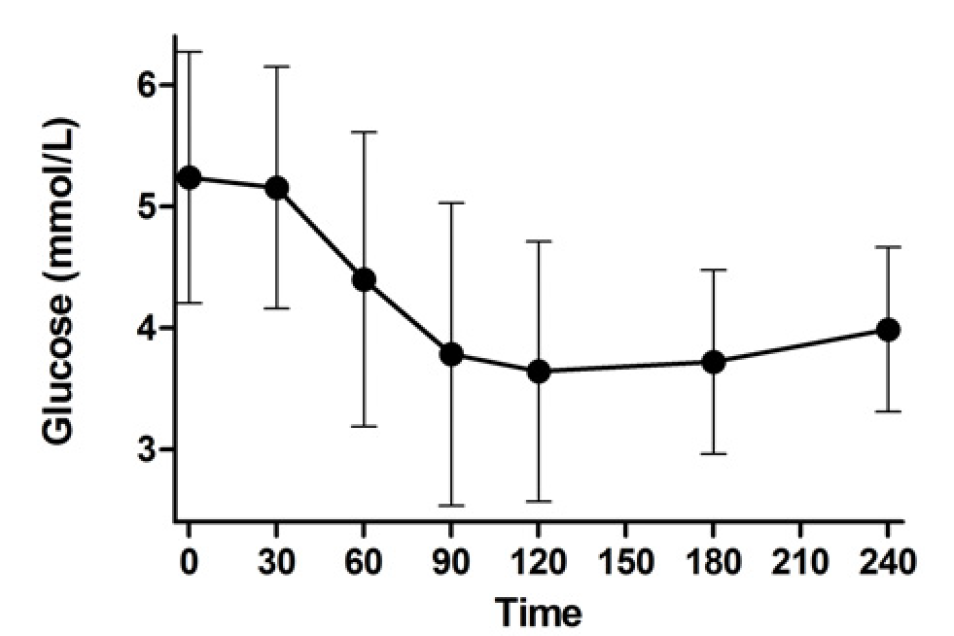
Ultimately, we wish for discovery to improve human health. Genomic inquiry may impact disease categorization, patient stratification, clinical prediction, drug discovery or therapeutic targeting. Genomic discovery that is agnostic to pre-existing knowledge has uncovered dozens of loci that influence glycemic dysregulation. Physiological investigation has begun to define disease subtypes, clarifying heterogeneity and suggesting molecular pathways for intervention. Convincing genetic associations have paved the way for the identification of effector transcripts that underlie the phenotype, and in select cases genetic or experimental proof of gain or loss of function has clarified the direction of effect to guide therapeutic development. Genetic studies can also examine off-target effects and furnish causal inference. By curating this information and making it widely available to all stakeholders, we hope to help enhance therapeutic development pipelines by accelerating efficiency, maximizing cost-effectiveness and raising ultimate success rates.
Pharmacogenetics in the Florez lab remains a key objective and approach, subdivided into three separate but related goals:
1) Patient stratification- genetic data may be used to categorize individuals into subgroups based on clinical response to the drug of interest.
2) Target identification- agnostic genome-wide studies may identify genes that encode drug targets, elucidating their mechanism of action and enabling the design of novel drugs that act on the same pathway.
3) Functional characterization- because drugs perturb the human organism in vivo, detecting a differential response based on a given genetic variant may illuminate the function of the gene produce encoded by the gene that harbors the variant or whose expression is influenced by it.
Thus the Florez lab uses drugs as handles that perturb the human organism in vivo. Whether in retrospective clinical databases (e.g. PHARMGen), prospective clinical trials (e.g. the Diabetes Prevention Program), or our newly designed physiological studies (e.g. SUGAR-MGH), we capture the human response to diabetes drugs and examine how genetic variation influences this response.
We hope to transform clinical practice. In our vision, once robust pharmacogenetic associations are discovered and confirmed, they will be aggregated into genetic risk scores that explain a substantial proportion of the variance in glycemic response or the appearance of side effects. They will be included into multi-trait genotyping arrays that capture all known clinically actionable genetic variants for most common diseases and approved drugs. This “megachip” would only need to be deployed once in the lifetime of an individual at an affordable cost, and the information could become part of that person’s electronic medical record. Statistical algorithms that define the likelihood of response could be created, tested, refined, and automatically triggered once a prescription order is initiated; decision support tools elaborated by experts would then inform the clinician, at the point of care, whether this specific patient is a good candidate for the selected agent. The methods and expertise exist to realize this vision: but it will not come to fruition unless we generate the required knowledge base, the data stand rigorous scrutiny, and cost-effectiveness analyses demonstrate that the benefit of clinical outcomes outweighs the expense incurred. We are invested in carrying out the studies needed to accomplish this goal.